CSV files
Introduction
rNAV is able to load a network from two CSV files. This chapter describe the CSV file format supported by rNAV.
Nodes file
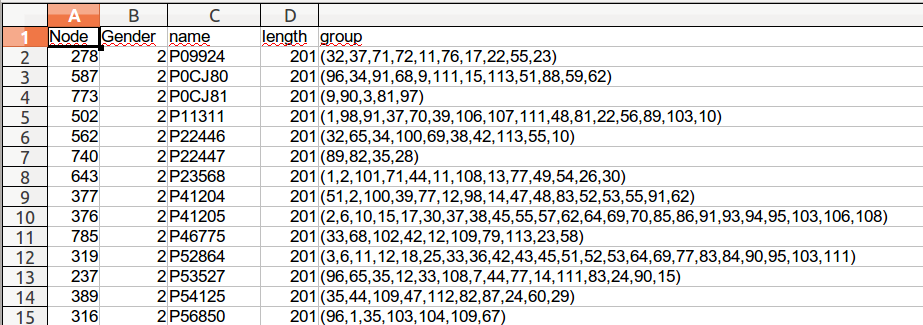
Nodes file must contain 5 columns:
Node: the node identifier, must be a unique integer
Gender: an integer
- 1 for mRNA
- 2 for sRNA
name:
- for mRNA, the UniprotKB gene identifier (if you want to use DAVID Chart enrichement tool), or any other name
- for sRNA, just its name
length: the length of the mRNA in nucleotide
group: list of neighbors identifier for each mRNA. And for sRNAs, just its own identifier.
Edges file
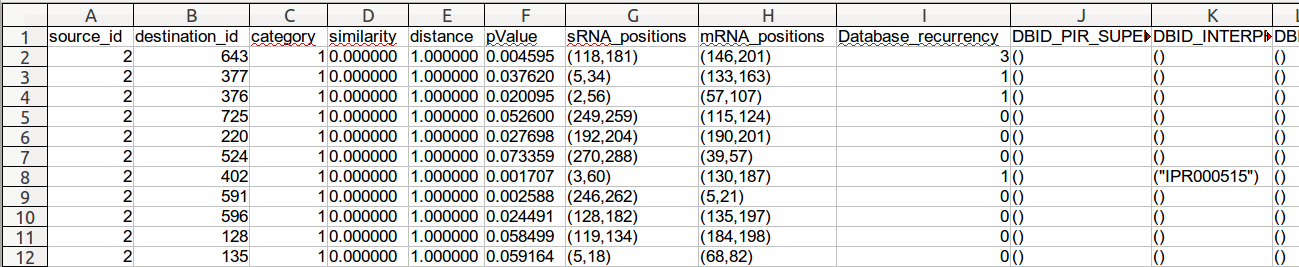
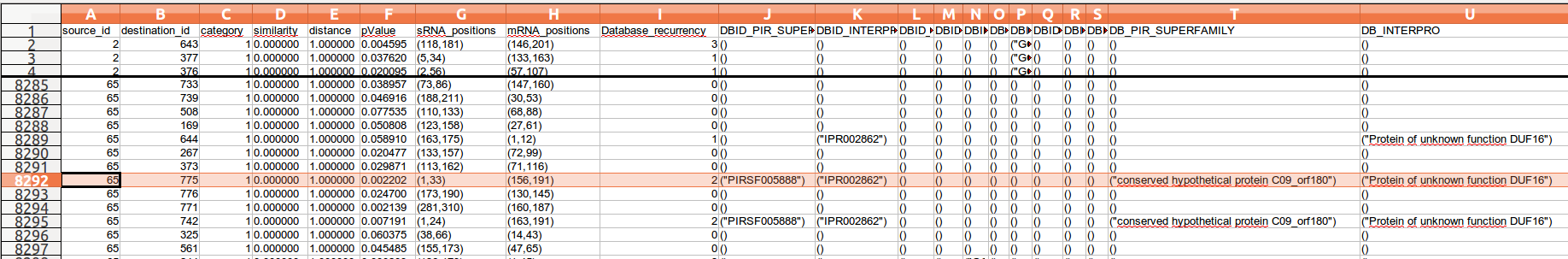
- source_id: the identifier of the source node of the edge
- destination_id: the identifier of the destination node of the edge
- category:
- similarity: the similarity score given by the alignement sequence program
- distance:
- pValue: the P-Value given by the alignement sequence program
- sRNA_positions: the position of the interaction defined by its start position and its end position on the sRNA (between brackets and commas separated)
- mRNA_positions: the position of the interaction defined by its start position and its end position on the mRNA (between brackets and commas separated)
- Database_recurrency: total number of annotations for this edge (all databases included)
- Database identifier & database content: the number of columns depends on the number of databases interrogated for annotation. For each database this file must content 2 columns with specific names.
- one column for database identifier named DBID_<DB_name>
- the second for database annotation terms named DB_<DB_name>